Hopefully all the vaccines I got will prevent severe COVID-19. I did get COVID-19 this summer during a trip to Mexico but it must have been the Omicron variety since I barely got sick at all.
Severe COVID-19 is associated with molecular signatures of aging in the human brain
Nature Aging (2022)
Abstract
As coronavirus disease 2019 (COVID-19) and aging are both accompanied by cognitive decline, we hypothesized that COVID-19 might lead to molecular signatures similar to aging. We performed whole-transcriptome analysis of the frontal cortex, a critical area for cognitive function, in individuals with COVID-19, age-matched and sex-matched uninfected controls, and uninfected individuals with intensive care unit/ventilator treatment. Our findings indicate that COVID-19 is associated with molecular signatures of brain aging and emphasize the value of neurological follow-up in recovered individuals.
Main
COVID-19 is an acute respiratory disease often accompanied by neurological sequelae1. Individuals with previous severe COVID-19 exhibit a 10-year average drop in their global cognitive performance2, mimicking accelerated aging. Complementary studies combining neuroimaging and cognitive screening implicate COVID-19-induced impairment of the frontal cortex3,4, a critical area for cognitive function, but molecular evidence of aging-like effects in the brain is lacking.
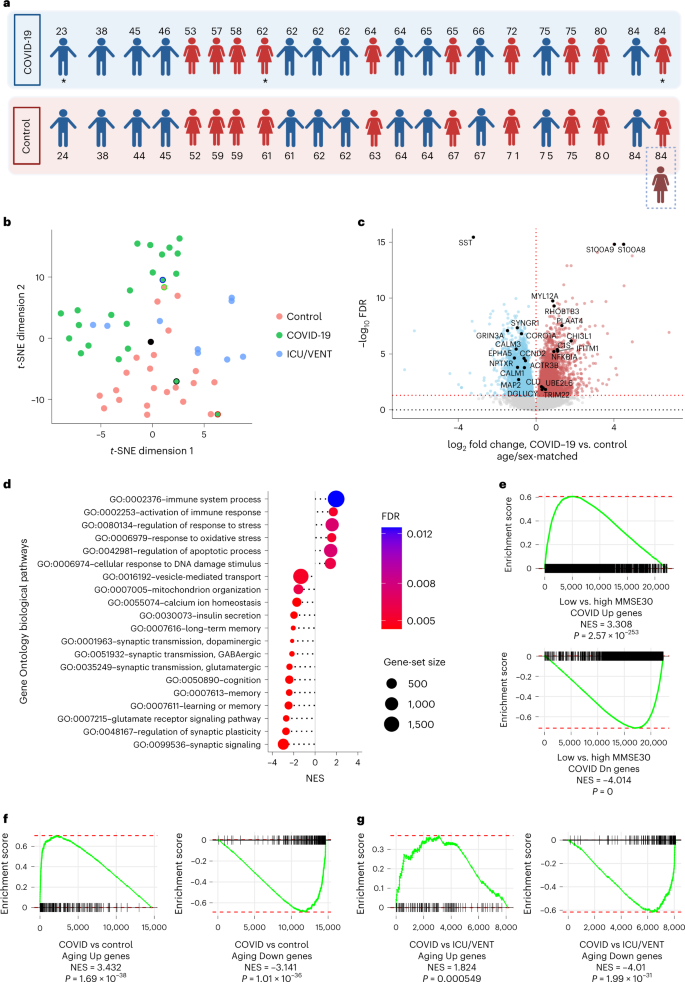
To address this, we performed RNA-sequencing (RNA-seq) analysis of 54 postmortem frontal cortex samples, including samples from 21 individuals with severe COVID-19 (previous neurological history was limited to Alzheimer’s disease in one person and epilepsy in another) and 1 asymptomatic individual aged between 23 and 84 years old, 22 age-matched (±2 years) and sex-matched uninfected controls with no history of neurological or psychiatric disorders, an age-matched and sex-matched uninfected individual with Alzheimer’s disease, and an additional independent control group of 9 uninfected individuals with history of intensive care unit (ICU) or ventilator treatment (22–85 years old; ICU/VENT; Fig. 1a and Supplementary Fig. 1a; see Supplementary Table 1 for clinical information; COVID-19 cohort). All COVID-19-cases were determined by positive pre-mortem or peri-mortem testing for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection via nasopharyngeal swab qPCR and history of hospitalization, whereas uninfected control samples were collected before the COVID-19 pandemic (with three exceptions in the ICU/VENT group that had negative SARS-CoV-2 qPCR tests at the time of death, and no COVID-19 history and/or negative serological test).
a, Age and sex of each individual in COVID-19 or uninfected age/sex-matched control (±2 years) groups (n = 22 per group) analyzed in this cohort. An asterisk indicates notable COVID-19 cases. The 23-year-old male presented with asymptomatic COVID-19, the 62-year-old female presented with severe COVID-19 history and comorbid epilepsy and the 84-year-old female who had a history of severe COVID-19 with comorbid Alzheimer’s disease (AD; an uninfected individual with AD was also included as an additional control; Supplementary Table 1). Created with BioRender.com. b, t-distributed stochastic neighbor embedding (t-SNE) analysis of frontal cortex transcriptomes from COVID-19-infected individuals, uninfected age-matched and sex-matched controls, and an independent group of uninfected controls with history of ICU and/or ventilator treatment (ICU/VENT). Black border, 23-year-old asymptomatic male with COVID-19. Red border, 62-year-old female with COVID-19 history and comorbid epilepsy. Blue border, 84-year-old female with COVID-19 history and comorbid AD. Black point, 84-year-old female without COVID-19 but with AD. Green border, uninfected age-matched and sex-matched control (non-AD) for the COVID-19-infected individual with comorbid AD. For age-matched/sex-matched controls and COVID-19 samples n = 22 per group; ICU/VENT-treated uninfected controls n = 9. c, Volcano plot representing the DEGs of the frontal cortex of individuals with COVID-19 versus age-matched and sex-matched controls (n = 22 per group). Red points denote significantly upregulated genes among COVID-19 cases (false discovery rate (FDR) < 0.05). Blue points denote significantly downregulated genes among COVID-19 cases. Black points highlight significant genes with corresponding gene symbols (Supplementary Table 2). d, Gene Ontology (GO) biological pathway enrichment analysis of COVID-19 versus age-matched/sex-matched control DEGs. Gene ranks were determined by signed −log10 FDRs of DEGs (Supplementary Table 3). e, GSEA of cognitive decline-regulated genes using COVID-19 (COVID-19 versus age-matched and sex-matched controls) DEGs. DEG ranks were assigned by signed −log10 FDR from the frontal cortex transcriptome of individuals with MMSE scores > 25 (high cognitive performance) versus the transcriptome of individuals with MMSE scores < 25 (low cognitive performance/cognitive decline) as measured in the ROSMAP study. f,g, GSEA of COVID-19 DEGs (COVID-19 versus age-matched/sex-matched control in f and COVID-19 versus ICU/VENT in g), using significantly upregulated (top) or downregulated genes (bottom) in our aging cohort as gene sets. DEG ranks were assigned by signed −log10 FDR from COVID-19 versus corresponding control frontal cortex. NES, normalized enrichment score. P, two-tailed GSEA P value (Supplementary Fig. 5).
By clustering analyses, COVID-19 transcriptomic cases broadly segregated away from controls, with two of the outliers being from the 23-year-old asymptomatic individual and the 62-year-old individual with comorbid epilepsy; the age-matched/sex-matched controls proximal to COVID-19 cases were from older adults (Fig. 1b and Supplementary Figs. 1 and 2). Uninfected older adults in the ICU/VENT group generally clustered closer to COVID-19-infected individuals, whereas younger ICU/VENT individuals clustered relatively close to controls; two cases cluster separately from all other samples (Supplementary Fig. 1b). Comparison of COVID-19 cases and their corresponding age-matched and sex-matched controls revealed 6,993 differentially expressed genes (DEGs), 3,330 of which were upregulated and 3,663 downregulated (Fig. 1c and Supplementary Table 2). For example, the S100A8 and S100A9 genes, which encode calprotectin and blood circulating levels of which distinguish severe from mild COVID-19 disease5, were upregulated among individuals with COVID-19. Pathway enrichment analysis identified numerous significant GO terms associated with aging in the human brain enriched upon severe COVID-19, including positive enrichment of immune-related pathways and negative enrichment of synaptic activity, cognition and memory pathways (Fig. 1d and Supplementary Fig. 3). We also observed significant associations of cellular response to DNA damage, mitochondrial function, regulation of response to stress and oxidative stress, vesicular transport, calcium homeostasis6, and insulin signaling/secretion7 pathways previously associated with aging processes and brain aging6,8. Altogether, our analyses suggest that many biological pathways that change with natural aging in the brain also change in severe COVID-19.
As natural brain aging is associated with cognitive decline, we further assessed associations of transcriptomic changes in COVID-19 and cognitive function. We collated frontal cortex transcriptomic data from 633 individuals who underwent the Mini-Mental State Examination (MMSE) while alive and donated their brains at the time of death as part of the ROSMAP study9,10. We split individuals and their corresponding transcriptomic data based on the median MMSE score: ≥25 as high cognitive performance versus MMSE < 25 as low cognitive performance. From gene-set enrichment analysis (GSEA), we found strong associations between low cognitive performance and COVID-19 (Fig. 1e).
Given the strong associations between aging-regulated pathways and severe COVID-19, we sought to directly test whether COVID-19 is associated with similar gene expression patterns as natural aging in the human brain. We performed RNA-seq analysis of postmortem frontal cortex samples of 10 young (≤38 years old) and 10 older (≥71 years old) uninfected individuals (Supplementary Table 1; aging cohort and Supplementary Fig. 4a) and compared these findings to COVID-19 DEGs. We found striking similarities between individuals with COVID-19 and aged individuals: genes upregulated in aging were upregulated in severe COVID-19; likewise, genes downregulated in aging were also downregulated in severe COVID-19 (see Fig. 1f for age-matched/sex-matched controls versus COVID-19). As further validation, we collated transcriptome-wide datasets from five independent aging cohorts and confirmed this association (Supplementary Fig. 5 and Supplementary Table 4). Intriguingly, we continued to observe a significant association between aging-associated genes and DEGs from individuals with COVID-19 versus uninfected individuals treated with ICU/VENT (Fig. 1g).
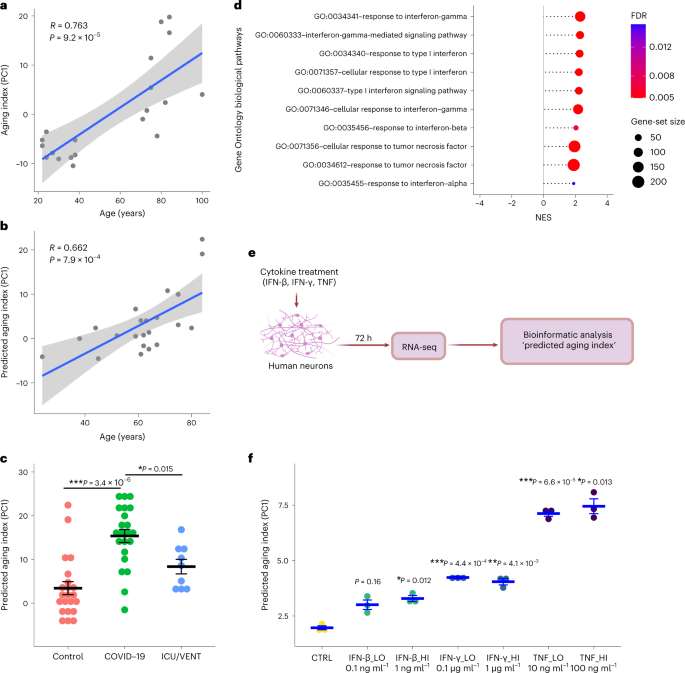
To delineate the effects of severe COVID-19 on brain aging directly, we leveraged our aging cohort to derive an aging index (Supplementary Fig. 4), comprising our aging DEGs and condensed by the first principal component across these transcriptomic data. As validation, we applied our predicted aging index to uninfected controls (COVID-19 cohort) and found similarly strong Pearson correlations between the training and test sets (Fig. 2a,b). Applying this model to individuals with COVID-19, we observed a significant increase in the predicted aging index compared to corresponding uninfected age-matched/sex-matched control and uninfected ICU/VENT control groups (Fig. 2c). Additional analysis revealed that the predicted aging index in individuals with COVID-19 was not significantly affected by the presence or absence of cerebrovascular disease (P > 0.05). Thus, severe COVID-19 appears to shift the molecular age of brains relative to both uninfected age-matched and sex-matched controls as well as uninfected ICU/VENT controls. Lastly, using qPCR analysis, we validated several of the top shared DEGs between our COVID-19 cohort and aging datasets (n = 22 per group; Supplementary Fig. 6).
a, First, a principal-component analysis using DEGs (FDR < 0.05) of young versus old uninfected controls estimated principal component 1 (PC1). The graph presents a two-tailed Pearson correlation of chronological age with aging index (PC1) among young versus old uninfected controls from the aging cohort (training set). n = 20. Gray shadow indicates the 95% confidence interval from a linear regression fit (R2 (18) = 0.58, P = 9.2 × 10−5). b, Two-tailed Pearson correlation of chronological age with predicted aging index (PC1) among uninfected age-matched and sex-matched controls from the COVID-19 cohort (test set). n = 22. Gray shadow indicates the 95% confidence interval from a linear regression fit (R2 (20) = 0.438, P = 7.9 × 10−4). c, Predicted aging index (PC1) of individuals with COVID-19 (n = 22), age-matched/sex-matched uninfected controls (control; n = 22), and an independent group of uninfected cases with ICU/VENT treatment history (n = 9). The line in each group represents the mean ± s.e.m. COVID-19 versus control Welch two-tailed t(42.0) = 5.68, P = 3.4 × 10−6; COVID-19 versus ICU/VENT Welch two-tailed t(21.0) = 3.14, P = 0.015. A Bonferroni correction was used to adjust for multiple comparisons. d, Significant interferon and TNF-related pathways identified using GO biological pathway enrichment analysis of COVID-19 versus age-matched/sex-matched control frontal cortex DEGs. FDR, GSEA FDR (Supplementary Table 3). e, Experimental design of in vitro cytokine treatment in human neurons. Created with BioRender.com. f, Effects of IFN-β, IFN-γ and TNF on predicted aging index, as assessed following in vitro treatment of primary human neurons. The line in each group represents the mean ± s.e.m. n = 3 independent wells (1 × 105 cells per well were plated) for each treatment. IFN-β_LO versus control Welch two-tailed t(2.68) = 4.48, P = 0.16; IFN-β_HI versus control Welch two-tailed t(3.51) = 8.26, P = 0.012; IFN-γ_LO versus control Welch two-tailed t(3.58) = 20.8, P = 4.4 × 10−4; IFN-γ_HI versus control Welch two-tailed t(3.35) = 12.3, P = 4.1 × 10−3; TNF_LO versus control Welch two-tailed t(3.63) = 33.6, P = 6.6 × 10−5; TNF_HI versus control Welch two-tailed t(2.28) = 15.8, P = 0.013. Bonferroni correction was used to adjust for multiple comparisons.
Finally, we sought to determine pathophysiologic mechanisms that may explain the association of COVID-19 with aging. We considered that this could be due to multiple factors, including SARS-CoV-2 viral infection in the frontal cortex or COVID-19-induced systemic inflammation. In agreement with previous studies11,12, SARS-CoV-2 viral RNA was not detected in samples from individuals with COVID-19 (at the time of death; Supplementary Fig. 7), suggesting that the observed gene expression changes are unlikely due to direct effects of the viral RNA in the frontal cortex. On the other hand, our transcriptomic pathway analyses identified upregulation of tumor necrosis factor (TNF) and type I/II interferon response pathways in the frontal cortex of individuals with COVID-19 (Fig. 2d). Indeed, interferons and TNF have been implicated in brain aging and aging-induced cognitive decline6,13,14,15. Among individuals with COVID-19 with available peripheral cytokine data, we indeed observed increased TNF levels (3.5–24.2 pg ml−1 at 0–2 d before death in three individuals; values > 2.8 pg ml−1 are considered elevated; data were not available for the rest of the individuals). In line with our findings, a mouse model of SARS-CoV-2 infection exhibited elevation of pro-inflammatory cytokines in cerebrospinal fluid, including interferon gamma (IFN-γ) and TNF, in the absence of viral neuroinvasion16. To test whether TNF and type I/II interferons can modulate the expression of aging-regulated genes, we performed a transcriptomic analysis (RNA-seq) on human primary neurons treated with different doses of TNF, interferon beta (IFN-β) or IFN-γ. Interestingly, TNF and to a lesser extent IFN-γ and IFN-β increased the predicted aging index of cytokine-treated neurons, suggestive of the induction of an aging-like effect (Fig. 2e,f). We also found that cytokines upregulated the expression of aging-regulated genes that were upregulated in individuals with COVID-19 such as TRIM22, CHI3L1, C1S and IFITM1 and downregulated the expression of aging-regulated genes that were downregulated in individuals with COVID-19 such as CCND2, ACTR3B and EPHA5 (Supplementary Fig. 8). Taken together, our data suggest that COVID-19-induced TNF and type I/II interferons may lead to significant deteriorating effects in the brain in the absence of SARS-CoV-2 neuroinvasion.
Aging is a major risk factor for the development of cognitive deficits. Our results, together with previously reported residual cognitive deficits reported in recovered cases2, imply that aging-associated and cognitive decline-associated gene expression changes observed in individuals with COVID-19 may lead to increased rates of cognitive decline. Furthermore, we provide evidence that these aging-regulated gene expression changes may be mediated in part by circulating TNF and type I/II interferons, suggesting that acute management of severe COVID-19-induced inflammation may be neuroprotective. We recognize limitations in our study including: the variability in illness duration, the imperfect quality of several samples and the specificity of our findings due to COVID-19. Despite these constraints, our study was sufficiently powered to identify substantial transcriptome-wide changes in individuals with COVID-19. In addition to being larger than previously reported COVID-19 brain transcriptome studies11,12,17, our COVID-19 cases were matched by age and sex to uninfected controls, enabling the identification of aging-associated gene expression signatures in our samples. Furthermore, we included ICU/VENT uninfected samples as an additional independent control group to distinguish COVID-19 from other comorbidities requiring ICU monitoring and/or ventilation. The generalizability of our results to individuals who had mild COVID-19 or who recovered from COVID-19 remains to be determined. Given our findings, we advocate for neurological follow-up of individuals who recovered from COVID-19 and suggest potential clinical value in modifying risk factors to reduce the risk or delay the development of aging-related neurological pathologies and cognitive decline18.
More at link.
No comments:
Post a Comment