Solve the primary problem of 100% recovery and you don't have to work on the secondary problem of depression.
33% survivor depression (28 posts to September 2016)
Identification of a miRNA–mRNA regulatory network for post-stroke depression: a machine-learning approach
- 1Faculty of Rehabilitation Science, Nanjing Normal University of Special Education, Nanjing, China
- 2Department of Neurosurgery, Suzhou Kowloon Hospital, Shanghai Jiao Tong University School of Medicine, Suzhou, Jiangsu, China
- 3Rehabilitation Medicine Center, The First Affiliated Hospital of Nanjing Medical University, Nanjing, China
- 4Department of Thoracic Surgery, Suzhou Kowloon Hospital, Shanghai Jiao Tong University School of Medicine, Suzhou, Jiangsu, China
Objective: The study aimed to explore the miRNA and mRNA biomarkers in post-stroke depression (PSD) and to develop a miRNA–mRNA regulatory network to reveal its potential pathogenesis.
Methods: The transcriptomic expression profile was obtained from the GEO database using the accession numbers GSE117064 (miRNAs, stroke vs. control) and GSE76826 [mRNAs, late-onset major depressive disorder (MDD) vs. control]. Differentially expressed miRNAs (DE-miRNAs) were identified in blood samples collected from stroke patients vs. control using the Linear Models for Microarray Data (LIMMA) package, while the weighted correlation network analysis (WGCNA) revealed co-expressed gene modules correlated with the subject group. The intersection between DE-miRNAs and miRNAs identified by WGCNA was defined as stroke-related miRNAs, whose target mRNAs were stroke-related genes with the prediction based on three databases (miRDB, miRTarBase, and TargetScan). Using the GSE76826 dataset, the differentially expressed genes (DEGs) were identified. Overlapped DEGs between stroke-related genes and DEGs in late-onset MDD were retrieved, and these were potential mRNA biomarkers in PSD. With the overlapped DEGs, three machine-learning methods were employed to identify gene signatures for PSD, which were established with the intersection of gene sets identified by each algorithm. Based on the gene signatures, the upstream miRNAs were predicted, and a miRNA–mRNA network was constructed.
Results: Using the GSE117064 dataset, we retrieved a total of 667 DE-miRNAs, which included 420 upregulated and 247 downregulated ones. Meanwhile, WGCNA identified two modules (blue and brown) that were significantly correlated with the subject group. A total of 117 stroke-related miRNAs were identified with the intersection of DE-miRNAs and WGCNA-related ones. Based on the miRNA-mRNA databases, we identified a list of 2,387 stroke-related genes, among which 99 DEGs in MDD were also embedded. Based on the 99 overlapped DEGs, we identified three gene signatures (SPATA2, ZNF208, and YTHDC1) using three machine-learning classifiers. Predictions of the three mRNAs highlight four miRNAs as follows: miR-6883-5p, miR-6873-3p, miR-4776-3p, and miR-6738-3p. Subsequently, a miRNA–mRNA network was developed.
Conclusion: The study highlighted gene signatures for PSD with three genes (SPATA2, ZNF208, and YTHDC1) and four upstream miRNAs (miR-6883-5p, miR-6873-3p, miR-4776-3p, and miR-6738-3p). These biomarkers could further our understanding of the pathogenesis of PSD.
 Huaide Qiu
Huaide Qiu Likui Shen2,
Likui Shen2,  Ying Shen
Ying Shen Yiming Mao
Yiming Mao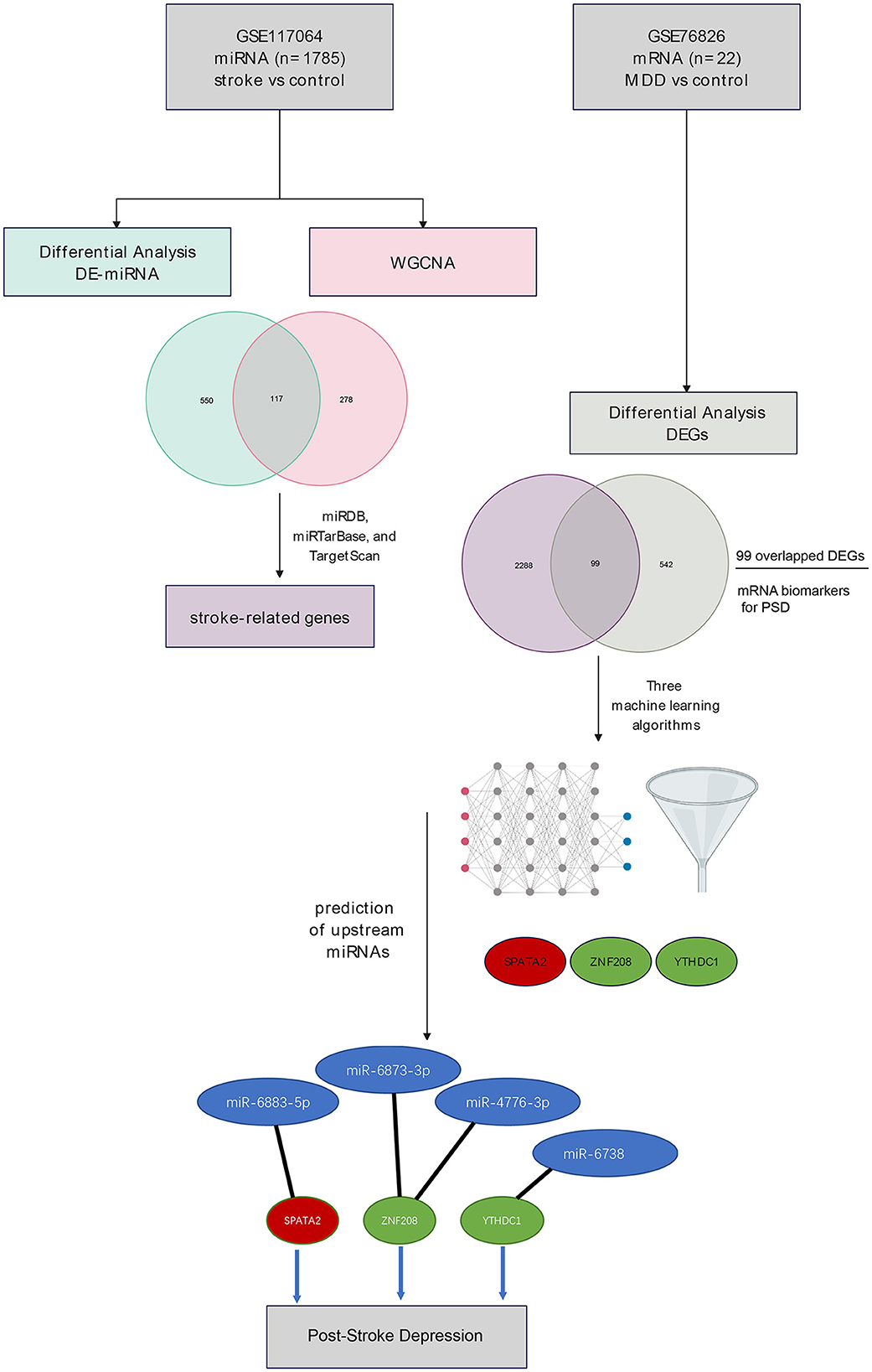
No comments:
Post a Comment