And yet even with a vast increase in speed you're still left with the neuronal cascade of death occurring in the first week. You do realize time-to-surgery is only the first step to 100% recovery? WHAT THE FUCK IS YOUR SOLUTION TO 100% RECOVERY after this intervention?
Deep Learning Assists with Stroke Evaluation and Management
R&E Foundation grant paves way for use of AI to accelerate time-to-surgery in stroke


According to the American Stroke Association, stroke is the leading cause of morbidity and fifth leading cause of mortality in the U.S., with an even greater disease burden worldwide.
To improve outcomes in patients with acute ischemic stroke, it is critical to quickly determine the need for endovascular surgery using CT angiography (CTA) of the head to assess for LVOs. However, rapid interpretation of CTA is difficult, particularly in settings without dedicated stroke centers and in areas where there are few radiologists.
“Because LVOs are extremely time-sensitive emergencies, an algorithm to identify them could be a gamechanger for patients with strokes by triaging cases that might be amenable to surgical treatment,” said Paul Yi, MD, director of the University of Maryland Medical Intelligent Imaging Center and assistant professor in the Department of Diagnostic Radiology and Nuclear Medicine at the University of Maryland School of Medicine in Baltimore.
Findings Provide Benchmarks for 2D vs. 3D Convolutional Neural Networks
With the support of a 2019 RSNA Research Resident Grant, Dr. Yi worked with colleagues, including a computer science team from Johns Hopkins University, to develop a high-performing deep learning system to detect LVO on CTA of the head.
The team used a dataset of 876 CTAs divided evenly between patients with and without LVOs. Each group had representation of both anterior and posterior circulation vessels.
They processed the dataset using standard neuroimaging procedures and reconstructed the images according to both axial and coronal views to correct for the issue that certain types of LVOs are more easily identified on one view or the other.
The processed datasets were then used to train, validate and test multiple deep learning algorithms using both 2D and 3D convolutional neural networks (CNNs).
According to Dr. Yi, the 2D CNN used heavily preprocessed images obtained through maximum intensity projection (MIP), a three-dimensional visualization process that can be used to display CTA data sets by converting 3D images into 2D images.
The 3D CNN used 3D stacks of images.
“Because CTAs are 3D volumes of images, we wanted to explore if 3D CNNs would provide an advantage over the 2D CNNs,” Dr. Yi said.
He added that he and his team wondered if pretraining the 3D CNNs on CTA-specific images, rather than general stroke images, would provide some advantage.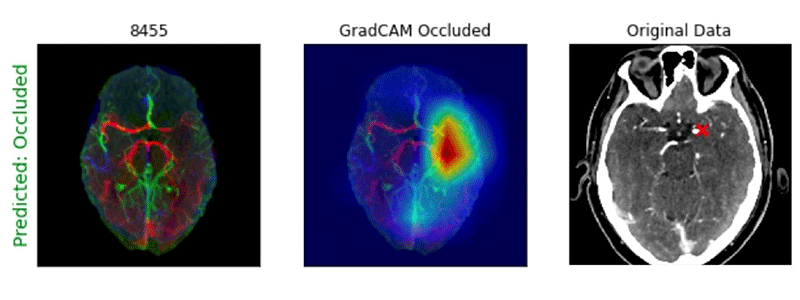
Example of Processed CTA Head image with GradCAM heatmap showing location of an M1 large vessel occlusion detected by a deep learning algorithm. Note that the “X” notes the site of the occlusion.
Image courtesy of Paul Yi, MD
The researchers found that overall, the 2D CNN using heavily preprocessed images performed best with an area under the curve (AUC) greater than 0.95, while the 3D CNNs achieved AUCs of 0.8 to 0.81 regardless of the pretraining method used.
In addition, the gradient-weighted class activation mapping tool (GradCAM) heatmaps, used for visualizing the parts of an image emphasized by a CNN model, were able to accurately localize LVOs for both the 2D and 3D CNN approaches.
Dr. Yi noted that from a methodological standpoint, the findings help provide benchmarks for using 2D versus 3D CNNs for volumetric medical imaging.
“While the 2D option may seem like the obvious choice because it performed better than 3D methods,” he said, “we stress that the 2D approach required heavy image preprocessing that may not be tenable in real-world clinical deployment.”
Dr. Yi noted that if further developed and clinically validated, the team’s work could help clinical radiologists in stroke care by triaging potential surgical emergencies.
“An algorithm like the one we developed could do a ‘pre-read’ of the study list, alert a radiologist to a scan with a potential emergency, and prioritize it for review,” he said.
Experience Affected by COVID, Infrastructure Challenges
As with many other researchers, Dr. Yi and his team conducted his research during the start and peak of the COVID-19 pandemic in spring 2020. Their normal in-person research activities were halted and moved remotely, however the computer-based nature of the work allowed them to continue their research.
In addition to the challenge of the pandemic, the team encountered a surprise in the foundational imaging informatics infrastructures they used to complete the project.
While identifying the study cohort and curating the images, Dr. Yi and his colleagues planned to perform batch extracts of images from their PACS. However, they found considerable variability in image storage and labeling conventions that made the automated identification of a relevant image series challenging.
In one example, he noted that each CTA had an average of 21 series, from which the team hoped to use only one series, the thin axial images. “This series ended up being labeled under 21 different names with 10% of the CTAs using multiple series with the same name,” Dr. Yi said.
Instead of automatic extraction, he and his team had to manually review the series for the correct inclusion. When they later shared their experience at the Society for Imaging Informatics in Medicine (SIIM) annual meeting in 2020, they learned other sites experienced the same difficulty.
“While discouraging in the moment, ultimately, I think this was a positive challenge because it helped shed light on an issue that is not an obvious consideration. It is definitely something I am now proactively addressing going forward for any project in the future,” Dr. Yi said.
“The grant helped me secure dedicated time to focus on my research and provided support for an engineering research scientist,” he said. “These were integral to my growth as a physician-scientist working in multidisciplinary research.”
PAUL YI, MD
No comments:
Post a Comment